Gene set enrichment analysis is an important tool linking results of statistical analyses of high throughput data with biological reality. The basic idea is that if you group genes into gene sets (for example, interferon stimulated genes) you will be able to answer questions such as: are interferon stimulated genes up-regulated in COVID-19? The R package tmod is a collection of algorithms, tools and visualisation methods for gene set enrichments.
What is special about tmod?
Firstly, tmod includes the CERNO algorithm for gene set enrichment analysis, which has been shown by us to have a high biological reproducibility and high sensitivity (Zyla et al. 2019). While similar in principle to the GSEA algorithm, it involves a statistic with a known distribution and thus does not require a randomization test strategy. This makes CERNO applicable in many situations in which GSEA is problematic.
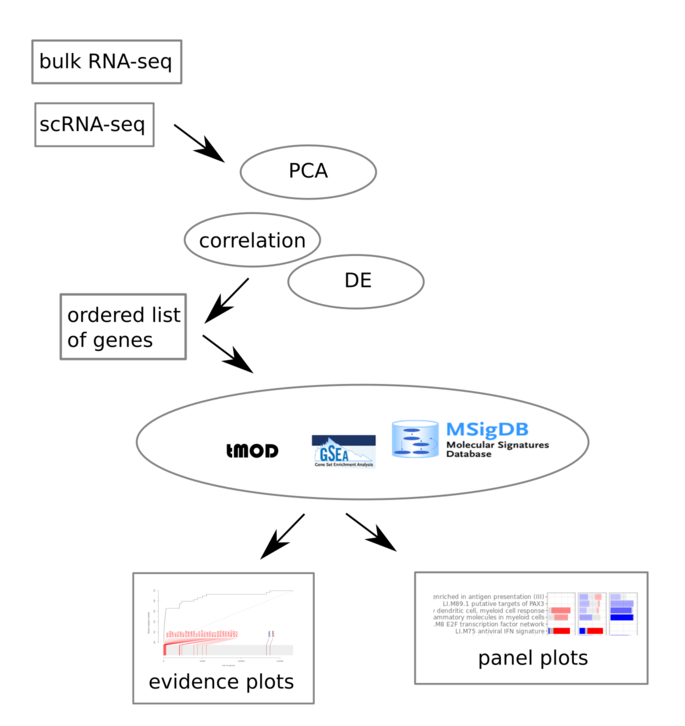
Secondly, tmod includes several gene sets based on transcriptional modules of human immune responses, which are not a part of any other software package. These are particularly suitable to study human diseases, even in animal models.
Thirdly, tmod includes new visualisation methods such as panel plots, which include both the information about effect sizes as well as p-values.
Finally, tmod integrates well with tidyverse.
Availability
CUBI Contact
Literature
- Zyla J, Marczyk M, Domaszewska T, Kaufmann SH, Polanska J, Weiner 3rd J. Gene set enrichment for reproducible science: comparison of CERNO and eight other algorithms. Bioinformatics. 2019 Dec 15;35(24):5146-54.
- Weiner 3rd J, Domaszewska T. tmod: an R package for general and multivariate enrichment analysis. PeerJ Preprints. 2016 Sep 4;4.
Last modified: Jan 3, 2023