Available automated methods for peak detection in untargeted metabolomics suffer from poor precision. We present NeatMS which uses machine learning to replace peak curation by human experts. We show how to integrate our open source module into different LC-MS analysis workflows and quantify its performance. NeatMS is designed to be suitable for large scale studies and improves the robustness of the final peak list.
Our the tool improves peak curation and contributes to the robust and scalable analysis of large-scale experiments. Furthermore, it provides all necessary functions to easily train new models or improve existing ones by transfer learning. NeatMS software is available as open source on github under permissive MIT license and is also provided as easy-to-install PyPi and Bioconda packages.
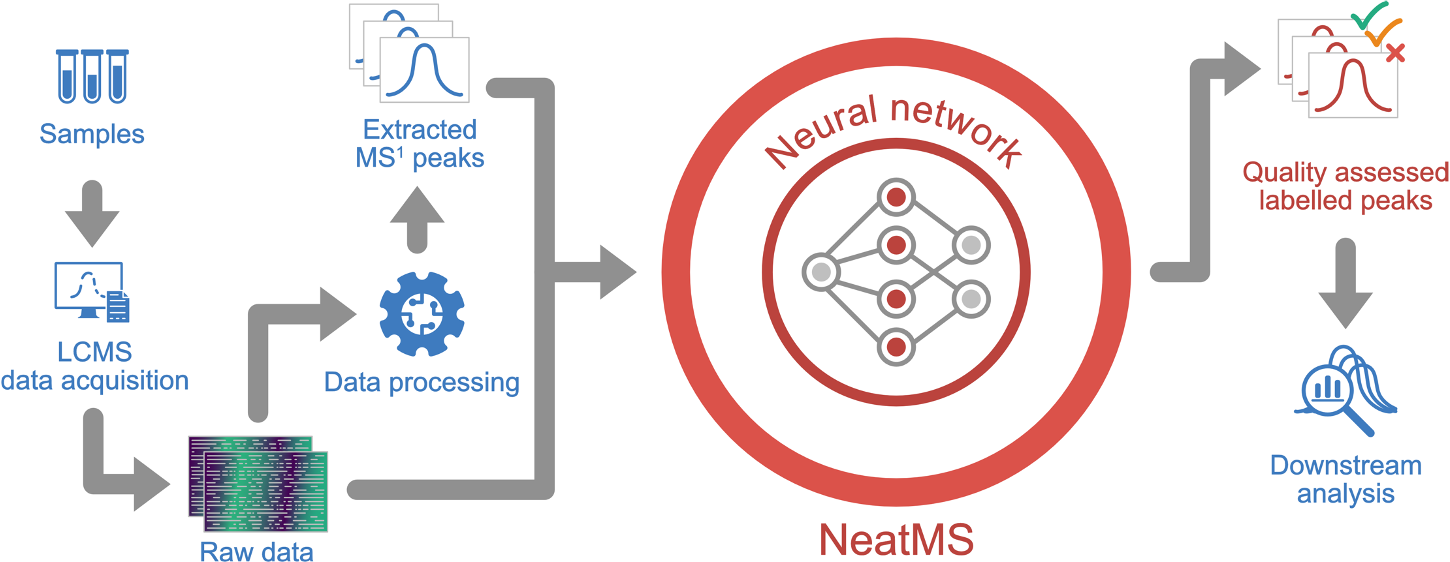
NeatMS Workflow
Publications
-
Gloaguen, Yoann, Jennifer A. Kirwan, and Dieter Beule. “Deep learning-assisted peak curation for large-scale LC-MS metabolomics." Analytical chemistry 94.12 (2022): 4930-4937.
-
Preprint on bioArxiv
Last modified: Jan 3, 2023