Targeted quantitative mass spectrometry metabolite profiling is the workhorse of metabolomics research. Robust and reproducible data are essential for confidence in analytical results and are particularly important with large-scale studies.
The increasing amounts of studies and data due to advancements in technology demands a shift toward automated and thus robust, reproducible, and reportable quality control. The metabolomics community has already recognized the need for more quality assurance and control in metabolomics studies. Community-wide collaborations such as the Metabolomics Quality Assurance and Quality Control Consortium (mQACC) further highlight the need and interest in quality control by creating a forum to discuss and implement community-wide procedures and standards for quality assurance and quality control. mQACC is providing education and developing standard reference material with focus on discovery-based (i.e., untargeted) methods. Our software complements these efforts with easy to apply measures for target studies in a readily usable, consistent, and reproducible manner. MeTaQuaC is available as an open source R package under a permissive MIT license. Source code, documentation, and issue tracking can be found at https://github.com/bihealth/metaquac.
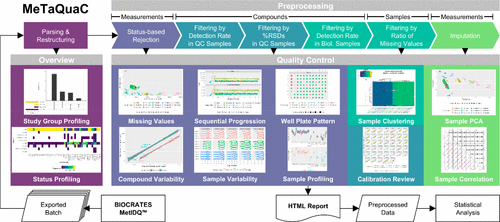
Visualisation of Quality Control Workflow
Related Publications
Kuhring, M.; Eisenberger, A.; Schmidt, V.; Kränkel, N.; Leistner, D. M.; Kirwan, J.; Beule, D. Concepts and Software Package for Efficient Quality Control in Targeted Metabolomics Studies – MeTaQuaC. Ana Chem 2020, https://doi.org/10.1021/acs.analchem.0c00136
Last modified: Mar 9, 2021