SODAR (System for Omics Data Access and Retrieval) has been developed by CUBI to provide easy and safe access to omics data, and its metadata, processing results, and logs. SODAR is a great help in organizing data and empowering users to manage access to their data and metadata curation. The main features of SODAR include web and command line interfaces for biomedical and computational users.
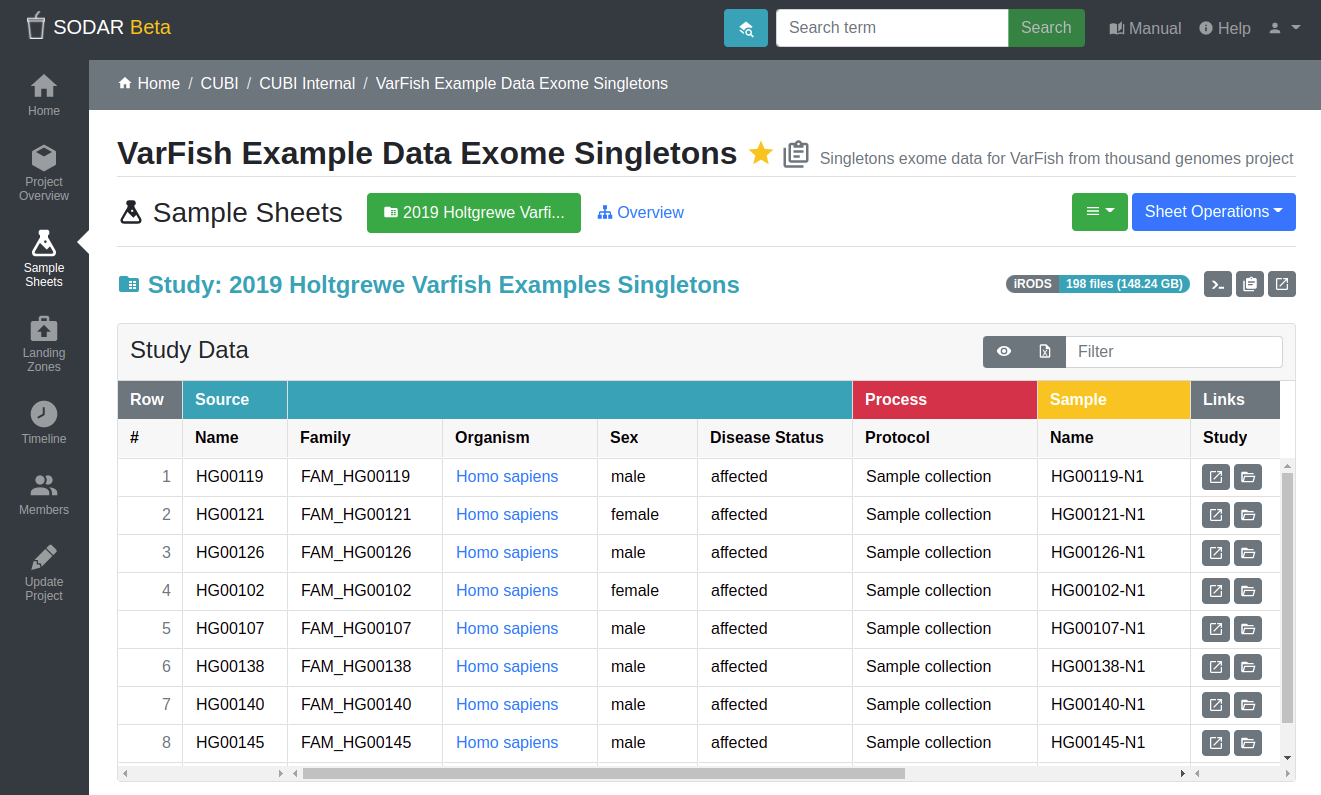
Access to studies in SODAR is controlled based on projects and PIs
Our local SODAR instance is reachable from internal networks only (Charite VPN access requires completing the form “Zusatzantrag B”. By default, data sets within SODAR are private. Access to a private project requires approval by the PI (or consortium) in charge of the data.
SODAR has been selected as the Omics data management solution for numerous large collaborative research projects including:
- Terminate-NB (Cancer Genomics)
- Translate NAMSE Berlin (Rare Disease Genetics)
- NUM-OrganoStrat (Single Cell Transcriptomics)
- BIH Clinical Single Cell Pipeline (Single Cell)
- MSTARS (Diverse Mass Spectrometry Technologies)
- SFB 1444 (Functional Geomics)
- Beyond the Exome (Genomics)
- BeLOVE (Multiomics)
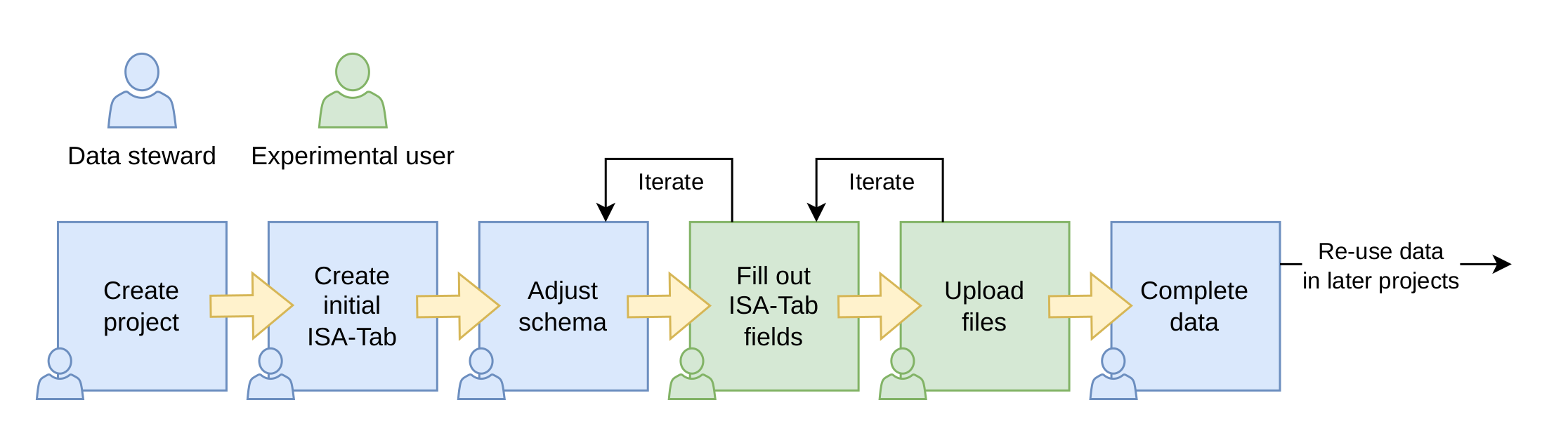
Meta-data flow in SODAR: the initial scheme is created by the data stewart (possibly a member of CUBI). The scheme is then iteratively improved by the person who generates the data, for example the experimental scientist.
Our local SODAR instance also hosts over 300 other projects, including smaller single PI ones. The total capacity provides petabytes of available storage space. SODAR follows the FAIR principles and uses community standards (Elixir) for metadata modelling.
User Documentation
- Introduction to SODAR – video by Mikko Nieminen
- Full SODAR reference
- SODAR demo page
Related CUBI Software
Last modified: Jan 4, 2023