CUBI is working closely together with the genomics and mass spectrometry (metabolomics and proteomics) platforms at MDC, Charite, and BIH. One area of this collaboration is managing and processing of the raw data. The initial use case is to support the genomics unit in data and meta data handling for Illumina sequencing data.
Sample Sheet Management & Demultiplexing
We have previously developed the web-based flow cell management platform DigestiFlow for which we also provide an in-house instance. Genomics facility staff is using this successfully for managing flow cells and sample sheets. In addition, DigestiFlow permits them to run the conversion from raw base calls (BCL) to sequence (FASTQ) files automatically.
DigestiFlow also automates the demultiplexing of sequenced library pools in an easy and reproducible fashion. Other features of DigestiFlow include the validation of sample sheets, comparing sample sheet adapter sequences with the sequenced bases in real-time, and warning about discrepancies. Finally, DigestiFlow creates a quality report for use by the genomics facility staff.
Data Delivery
We are currently developing the RODEOS system which will automate raw data management and data delivery from omics core units to users across BIH, Charite and MDC.
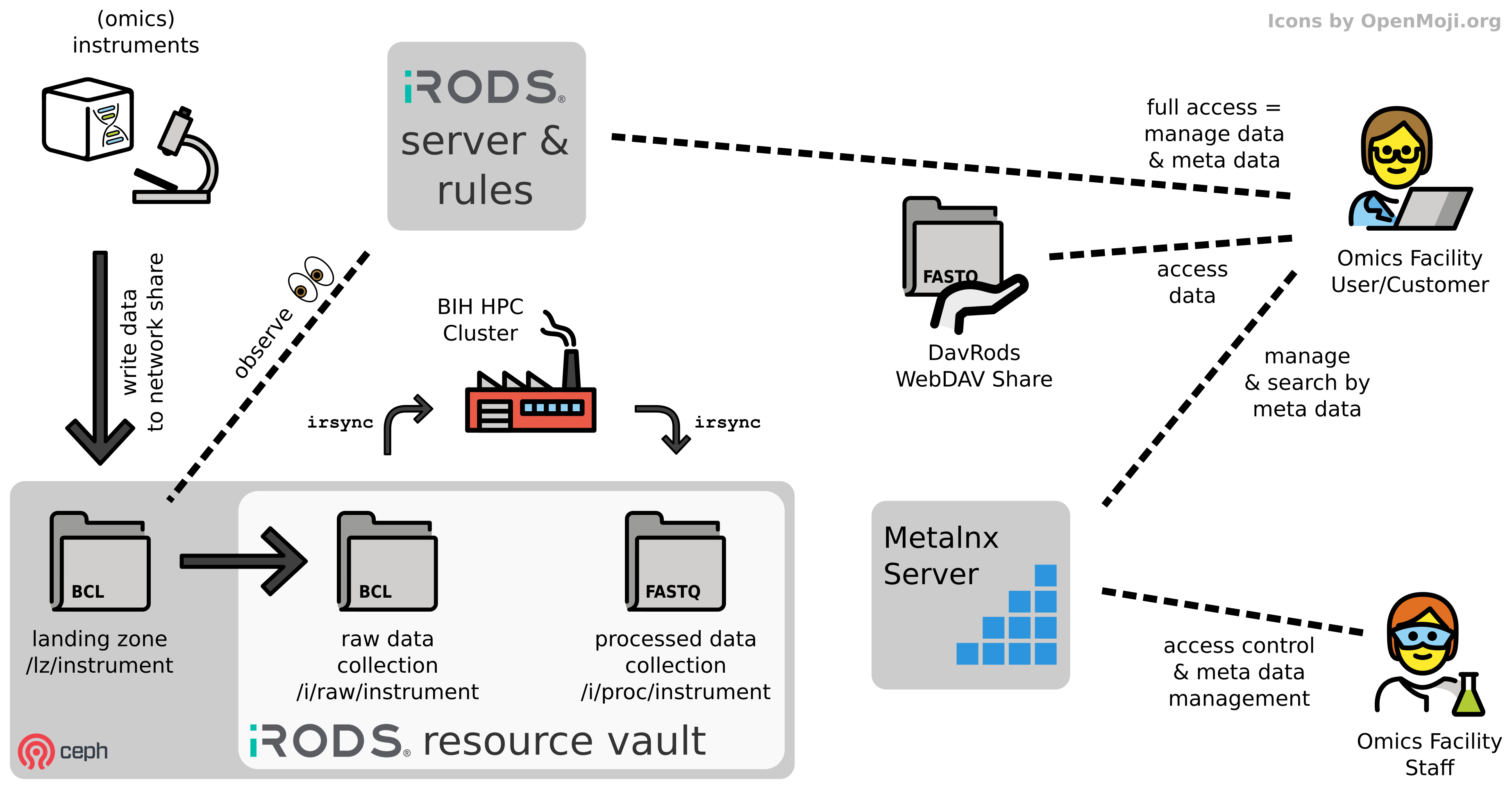
Related CUBI Software
Related CUBI Publications
More information on DigestiFlow can be found here on our website and the corresponding publication:
- Holtgrewe, M.; Messerschmidt, C.; Nieminen, M.; Beule, D. Digestiflow: From BCL to FASTQ with Ease. Bioinformatics 2019, btz850. doi:10.1093/bioinformatics/btz850.
Last modified: Mar 15, 2021